contents area
Public Health Weekly Report
detail content area
- Date2019-08-14 21:13
- Update2019-11-19 18:45
- DivisionDivision of Bacterial Diseases
- Tel043-719-7053
Analysis of surveillance results of pathogens from acute pneumoniae patients in children
Jung Sang Oun, Kim Sohyeon, Kim Hwanhee, Lee Chae Young, Yoo Jaeil, Hwang Kyuhjam
Division of Bacterial Diseases, Center for Laboratory Control of Infectious Disease, KCDC
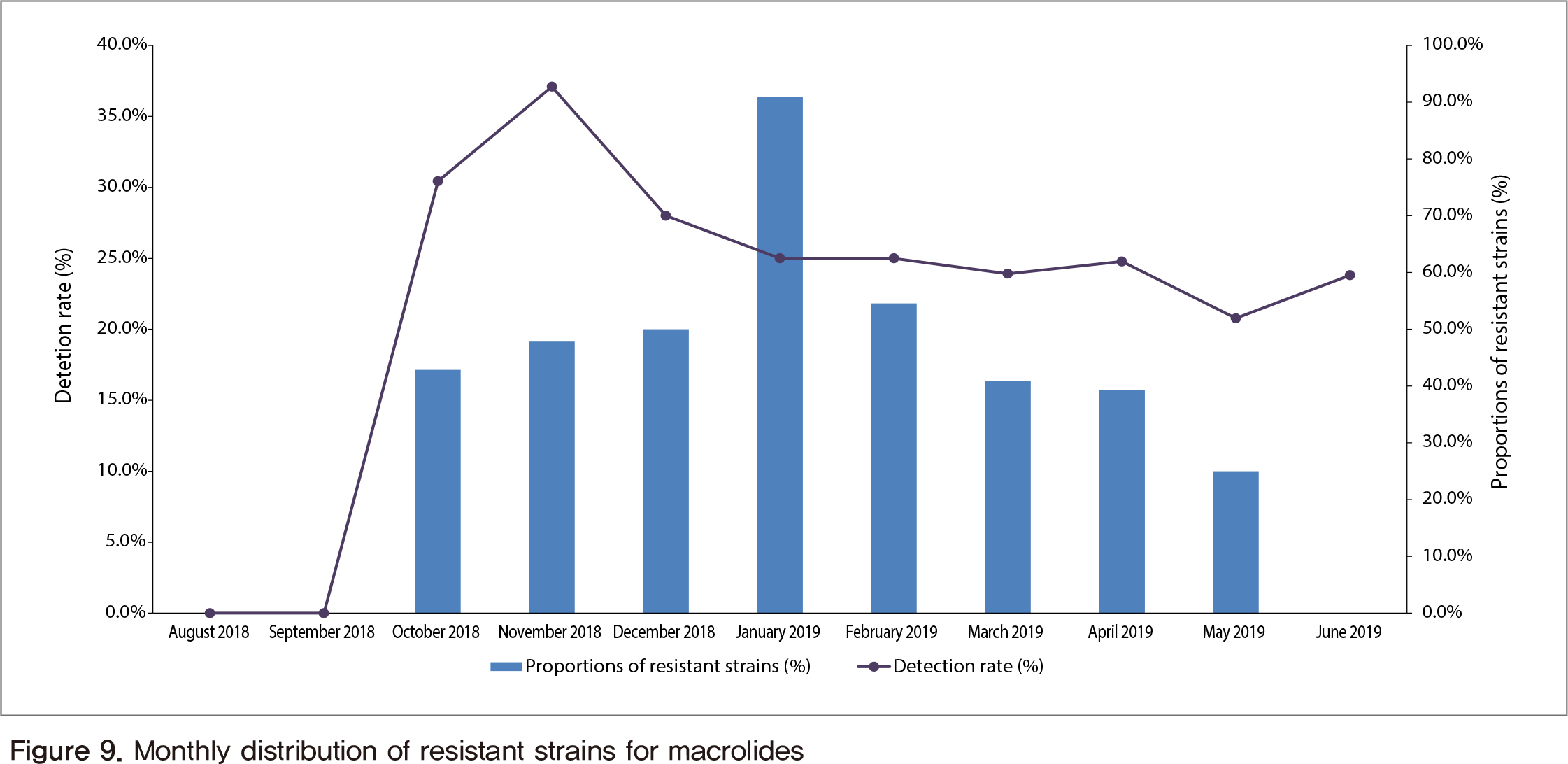
To know distribution status of pathogen in pediatric pneumonia and to confirm resistance rate for antibiotics of isolated bacterial pathogen, we had constructed hospital-based surveillance network with 27 regional cooperative hospitals, collected specimens from pneumonia patients in child with clinical signs, and performed laboratory tests for detection of bacterial/viral pathogen. If bacterial pathogens were isolated, their drug resistant patterns were confirmed by antibiotics susceptibility test. As the results, pathogens were identified from 92.2% of the collected specimens. The confirmed rate of viral pathogen (74.8%) was slightly higher than bacterial pathogen (55.6%). In the case of bacteria. Mycoplasma pneumoniae showed most high detection rate, followed by Staphylococcus aureus and Streptococcus pneumoniae. In virus, the highest one was human rhinovirus, followed by respiratory syncytial virus, metapneumovirus, adenovirus, bocavirus, parainfluenza virus, influenza virus, and coronavirus. In the analysis of monthly distribution status, positive rate of M. pneumoniae and S. pneumoniae increased in November 2018 and April 2019. Haemophilus influenza showed increased positive rate in Arpil and May 2019. In the case of viral pathogens, respiratory syncytial virus was detected high in November and December 2018. Then detection frequency of human rhinovirus was increased from February to May 2019. Metapneumovirus and parainfluenza virus also slightly increased in April 2019. Drug resistant pattern in bacterial pathogens, there was no resistant strain in Pseudomonas aeruginosa. However, resistance in ampicillin and amoxicillin/clavulanate was confiremd in H. influenzae. S. pneumoniae also showed high resistance in erythromycin, azithromycin, cefaclor and cefuroxime. High resistance in ampicillin and cefazolin was observed in Klebsiella pneumoniae and also high resistance in ampicillin was confirmed in S. aures. In the case of M. pneumoniae, the ratio of genetic variant type in 23S rRNA gene related to macrolide resistant was confirmed as 50.4%. As the conclusion, the analysis results obtained from this pathogen surveillance study in pediatric pneumonia will be used as a scientific basis for policy making of management of pediatric respiratory diseases.
Keywords: Pediatric pneumonia, Bacteria, Virus, Pathogen distribution, Antibiotics resistance







 This public work may be used under the terms of the public interest source + commercial use prohibition + nonrepudiation conditions
This public work may be used under the terms of the public interest source + commercial use prohibition + nonrepudiation conditions